Home> News
New research confirms the feasibility of retron editing in mammal cells
Updated: 2021-11-30
It is of great importance for the research of life sciences, disease diagnosis and treatment and the development of the agricultural sector to conduct precise modification of DNA sequences with high efficiency. The introduction of the CRISPR-Cas system has introduced greater precision to genome editing techniques.
The incorporation of donor DNA sequencing information into genomic DNA can be achieved when CRISPR-Cas9 occasions DNA double strand break (DSB) activities and then expedites insertion or indel in target loci through both non-homologous end joining (NHEJ) and homology-directed repair (HDR).
Theoretically speaking, it is plausible to precisely incorporate the information of DNA sequences into genomes to the maximum via HDR, though it is NHEJ rather than HDR that is responsible for repairing DSB under the mediation of CRISPR-Cas9. Research shows that utilizing single-stranded DNA as a template and increasing the concentration of DNA templates relative to Cas9-mediated DSB loci can effectively enhance the efficiency of HDR.
Bacteria have developed many specially designed defense systems like CRISPR in their own battle against phages. Recent studies show that retron systems have also played an important role in bacterial phage defense.
Their genetic elements are composed of a specialized reverse transcriptase (RT) and a relevant non-coding RNA (ncRNA) which can be partially reverse transcribed by RT initiating at a conserved guanosine (G) residue to produce a multicopy single-stranded DNA (msDNA).
Early in 2018, a research team led by Professor Hunter Fraser at Harvard University successfully designed CRISPEY (cas9-retron precise parallel editing with homology), which connects retron systems with CRISPR.
They found that after being reverse transcribed in cell through a retron system, the single-stranded donor DNA templates can be covalently tethered to sgRNA. By co-expressing Cas9 protein, reverse transcribed retron and retron ncRNA-sgRNA, which contain the sequences of donor templates, in yeast cells, the single-stranded donor DNA templates will be induced to approach Cas9-mediated DSB loci through sgRNA and Cas9.
This method can effectively allow for precise genome editing and enable the editing efficiency to reach as high as over 80 percent. Further research shows that this technique can produce a NDA sequence with 700bp in yeast cells in an efficient and precise manner. However, research on retron systems in mammal cell has not yet been conducted.
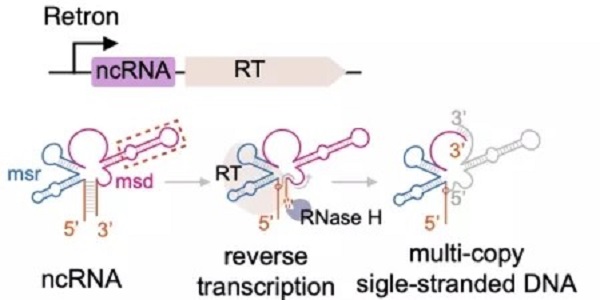
Composition of bacterial retrons and the birth of msDNA
A research team at Shanghai Huigene Therapeutics and the Yang Hui study group at the Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences jointly published a research paper titled "Precise Genome Editing without Exogenous Donor DNA via Retron Editing System in Human Cells" in the journal Protein & Cell on Aug 17.
By further optimizing retron systems, they confirmed the feasibility of applying them in mammal cells. Their laboratory studies will provide an important reference for the potential application of retron systems in higher-eukaryote cells.
By utilizing the expression of four reported functional retrons (Ec48 RT, Ec73 RT, Ec86 RT and Ec107 RT) in mammal cells, researchers thus confirmed that single-stranded DNA can be reverse transcribed through bacterial retrons in mammal species.
Coupled with 3′ Ec73 rgRNA, Cas9-Ec73 RT fusion can be harnessed for efficient and precise genome editing in mammal cells, boosting editing efficiency by 10 percent when retron RT is tethered to either RT-Cas9 or Cas9-RT and retron ncRNA is fused to either 5’ rgRNA or 3’ rgRNA.
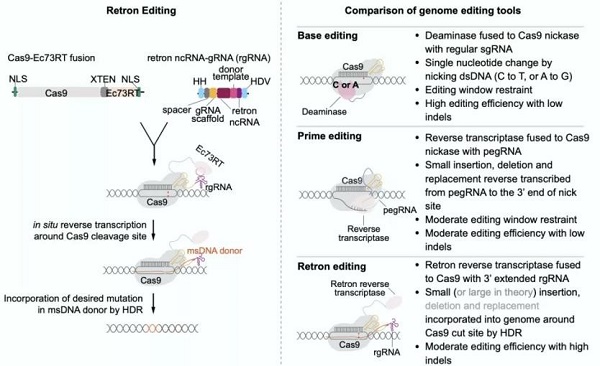
A work diagram of retron editing and its comparison with DNA base editing and prime editing
The researchers have conducted initial exploration of the application of retron editing techniques in mammal cells. To be noted, the research team led by Hunter Fraser, who have already implemented CRISPEY systems in yeast cells, have incorporated their previous studies on the utilization of retron systems into precise and efficient genome editing in mammal cells in an academic paper to be published online by BioRxiv.
Existing scientific findings have indicated the feasibility of integrating retron editing with more precise and efficient genetic editing in mammal cells. Given the relatively lower editing efficiency of current retron editing techniques and the high frequency of Cas9-induced indel, there are more strategies available to further renew and upgrade existing versions of the retron editing system.
For instance, an analysis of metagenomic data can uncover a highly efficient retron editing system. The improvement and upgrading of proteins will further enhance the activity of reverse transcriptase in existing retron RT. In addition, replacing Cas9 with Cas9 nickase will reduce the occurrence of negative by-products induced by DSB.
Compared with existing popular editing methods such as DNA base editing and priming editing, HDR-dependent retron editing features relatively lower restrictions from PAM loci. Meanwhile, retron editing systems don’t need additional exogenous donor DNA.
Therefore, a retron editing system might be a better option for genetic editing in plants because of the lower efficiency of HDR in plant cells and relatively low frequency of gene knock-in induced by the difficulties arising from delivering exogenous homologous recombination donor DNA.
Retron editing has natural strengths in terms of improving genetic editing. There is more work to be done to improve and upgrade it in order to facilitate its application in disease diagnosis and treatment and the improvement of pharmaceutical functions.
The research was co-conducted by Professor Kong Xiangfeng with Shanghai Huigene Therapeutics, as well as Professor Wang Zikang and graduate student Zhang Renxian at the Center for Excellence in Brain Science and Intelligence Technology under the guidance of Yang Hui.
Other contributors to the research included professors Shi Linyu and Wang Xing at Shanghai Huigene Therapeutics, and Zhou Yingsi at the Center for Excellence in Brain Science and Intelligence Technology.
